Categories Products for NMR Oligo Synthesis Service Uniformly labelled RNA Synthesis
Uniformly labelled RNA Synthesis
Uniformly Stable Isotope-Labelled RNA Oligonucleotide Synthesis Service
Dependent on isotope labelling specification and sequence length, Silantes has developed different RNA synthesis strategies. For synthesis of uniformly stable isotope-labelled (2H, 13C, 15N, 18O, 19F) RNA oligonucleotides, Silantes uses the T7 RNA polymerase system for in vitro transcription.
Uniformly labelled RNA oligonucleotides at a reasonable price
The synthesis costs for stable isotope-labelled RNA oligonucleotides are mainly driven by two factors: Stable isotope-labelled rNTPs raw materials and complex purification labor costs.
In order to make stable isotope-labelled RNA reagents affordable, Silantes has optimized the production process:
- Reasonably priced raw materials: the isotope-labelled rNTPs are made in-house by the company.
- Low purification costs: efficient synthesis using the T7 RNA polymerase system.
To save costs: The Silantes Feasibility Study
Silantes has a large and growing database of RNA sequences that have already been synthesized. Although RNA synthesis with the T7 RNA Polymerase System has been very successful, there are cases where the synthesis of the desired RNA product was challenging. To minimize the risk of synthesis projects for our customers, we offer the possibility of a prior multi-step feasibility study:
In the first step, the target RNA sequence is validated against Silantes' internal database. Then the target RNA sequence is screened for potential 3D structures that may complicate synthesis or purification. In the third step, to estimate yields, synthesis of the target RNA sequence is performed on an analytical scale with unlabelled rNTPs.
The project is terminated in case any of the above steps were unsuccessful and only the expenses for the feasibility study will be charged.
If the feasibility study is successful, the synthesis of the target RNA sequence is carried out on a preparative scale using isotope-labelled rNTPs. In this case, no costs for the feasibility study will be charged.
Silantes Method I: DNA template as the basis for transcription
T7 is a bacteriophage that produces its own RNA polymerase, which in turn uses its own promoter sequence as a start signal. This T7 promoter has the advantage that it results in very efficient RNA transcription and is not recognized by other RNA polymerases.
Silantes has optimized in vitro transcription with the T7 RNA Polymerase Sytstem and produces highly pure RNA sequences in only 3 steps which are outlined below.
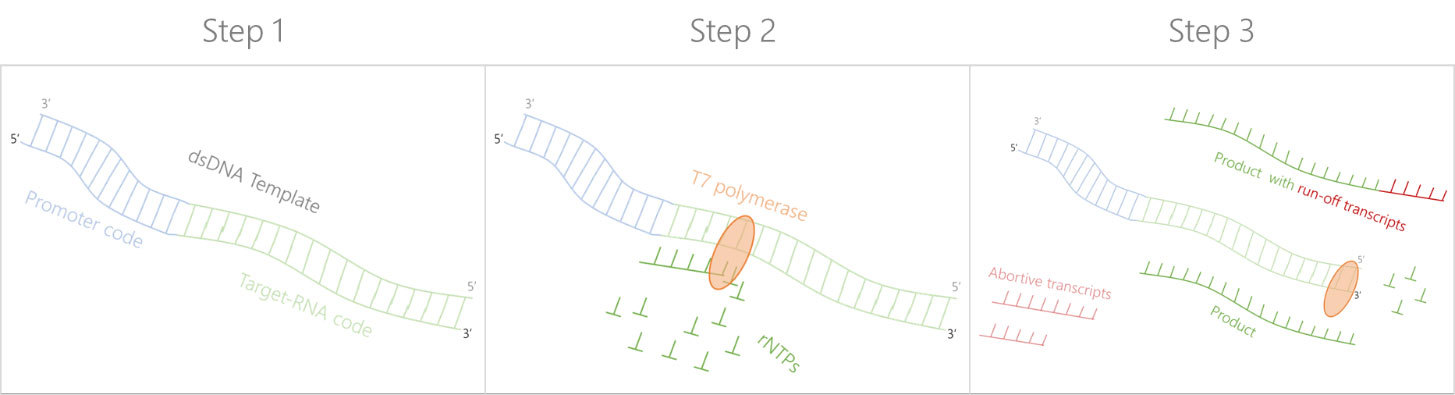
Step 1: DNA template preparation
Two complementary DNA single strands are synthesised, both containing the T7 promoter and the target RNA sequence. The two DNA single strands are then annealed. The DNA template forms the basis of transcription for the T7 polymerase.
Step 2: RNA transcription
The DNA template is mixed with the T7 polyermase and Silantes' isotope-labelled rNTPs. The T7 polyermase recognizes the promoter sequence within the DNA template and transcribes the isotope-labelled target RNA sequence.
Step 3: RNA purification
After transcription, the mixture made of isotope-labelled rNTPs, abortive transcripts, isotope-labelled target RNA sequence and run-off transcripts are fractionated by HPLC to obtain pure isotope-labelled target RNA.
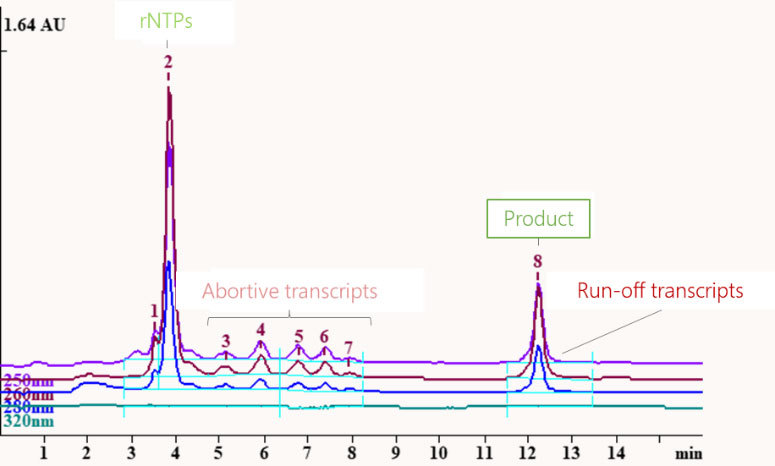
Limitation of the DNA template as transcription basis
Longer target RNA sequences require longer DNA templates. DNA templates are usually synthesized using solid-phase synthesis. However, the efficiency and quality of solid-phase synthesis decreases significantly as the length of the product increases.
T7 polymerase sometimes adds up to five or six nucleotides at the 3'-end of RNA transcripts. The longer the target RNA sequence, the smaller is the relative difference in length between target RNA sequence and run-off transcripts. Due to the small difference in length, run-off transcripts and target RNA are difficult to separate.2 To keep the cost of purification low, at Silantes synthesizes longer RNA sequences using the T7 polymerase system with ribozyme.
Silantes Method II: Plasmid as the basis for transcription
For long RNA sequences, Silantes uses a linearised plasmid instead of the DNA double-stranded template as the basis for transcription. Unlike long chemically synthesised DNA templates, long plasmids can be amplified in bacteria in a cost-saving and efficient manner.
The linearised plasmid used by Silantes contains information the T7 promoter and target RNA sequences. When the T7 polymerase transcribes the ribozyme at the 3' end of the plasmid, the ribozyme specifically cuts the product-ribozyme complex between the product and the ribozyme.3
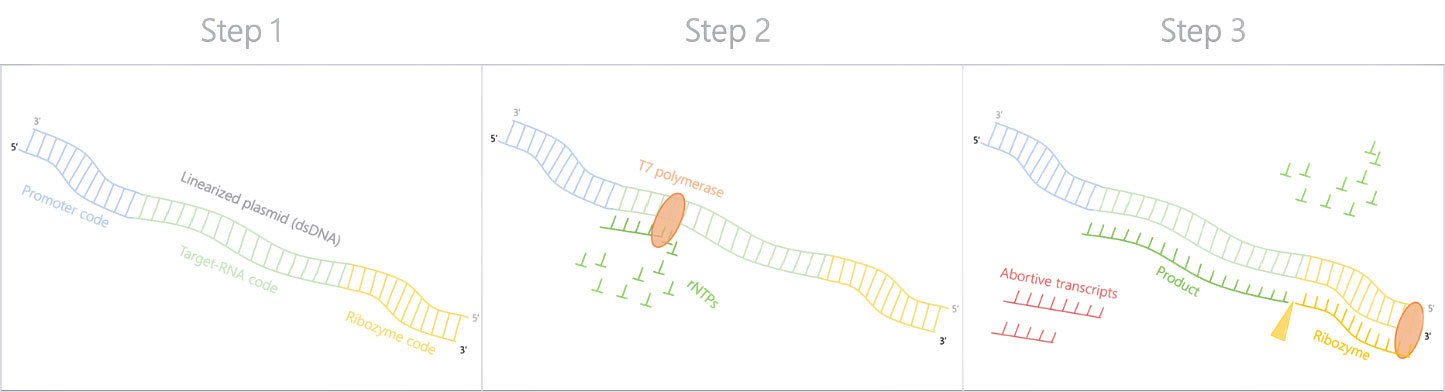
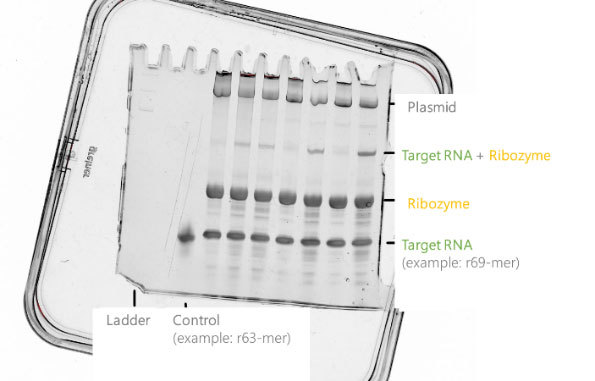
The in vitro synthesis of long RNA sequences using the T7 polymerase system with ribozyme results in very homogeneous 3'-ends of the target RNA sequence. In contrast to the synthesis with a DNA template, no complex and cost intensive purification of run-off transcripts is required. The SDS gel in Figure 9 shows a prominent band of the target RNA sequence.
Realize your RNA project with Silantes
If you need a stable isotope-labelled RNA sequence for your project, Silantes can take care of the synthesis work for you. For a quote we need the following information:
- Sequence of the target RNA (5’-GG end is required for RNAs. This can be appended to the RNA sequence if necessary)
- Desired isotope labelling (2H, 13C, 15N, 18O, 19F, or combinations thereof) and specification of isotope-labelling (entire oligonucleotide or all positions of a specific base)
- Desired quantity of target RNA
- Is purification desired? Depending on the sequence, Silantes experts will decide which method of purification (gel or HPLC) is best suited for your RNA.
Based on this information, we provide a quotation for the synthesis of your RNA fragment. Once an order is placed, purified RNA fragment delivery time is commonly less than 6 weeks.
For quotation inquiries or to get more information please contact sales@silantes.com.
2 Gallo, S., Furler, M. and Sigel, R. K. (2005) “In vitro Transcription and Purification of RNAs of Different Size”, CHIMIA, 59(11), p. 812. doi: 10.2533/000942905777675589.
3 Gallo, S., Furler, M. and Sigel, R. K. (2005) “In vitro Transcription and Purification of RNAs of Different Size”, CHIMIA, 59(11), p. 812. doi: 10.2533/000942905777675589

